Ja, tukaj gor od parih posebnežev, da so jedli konjske tablete proti zajedalcem in da je vse zarota...Garfildd
Me čudi da se od 2020 nisi prav ničesar naučil o maskah.
Za viruse mora biti ffp2 ali še raje ffp3.
V biolabih, kjer delajo z virusi ne boš videl prav nobenega, da bi imel kirurško masko gor.
Jabolka in kombajni.
Pri operacijah kirurgi zato, da ne pljuvajo svojih bakterij iz ust v odprte zadeve pacienta na mizi.
Pfizer doing SARS-COV2 Gain of Function
- Ustvarjalec teme AndY1
- Začetni datum
Uporabljate neposodobljen brskalnik. Ta ali druga spletna stran morda ne bo prikazana pravilno.
Posodobite brskalnik ali uporabite alternativni brskalnik.
Posodobite brskalnik ali uporabite alternativni brskalnik.
Aha, torej maske so čisto brez veze, čisto nič ne zmanjšajo možnosti prenosa virusov?



Malo ti še manjka, sej bo, Spet primerjava na nivoju vrtca.Predlagam ti, da se ne boš počutil zvezanega in prisiljenega v nekaj, da iz avta odmontiraš varnostne pasove in airbage,
saj večina jih ne rabi, tisti mali procent ki pa rešijo življenje je pa itak zanemarljiv...
Drugače pa airbage se da izključit, varnostni pas pa pač ne pripneš, ne rabiš nič šraufat, samo imaš možnost izbire. Ampak kaj ma to veze z razlikovanjem med večjim in manjšim, katero ti ne gre? A ti razumeš da z zajemanjem vode s cedilom ne moreš natankat posode ali ne?
Najbrž ne. Pa še en cajt ne boš.


Malo ti še manjka, sej bo, Spet primerjava na nivoju vrtca.
Drugače pa airbage se da izključit, varnostni pas pa pač ne pripneš, ne rabiš nič šraufat, samo imaš možnost izbire. Ampak kaj ma to veze z razlikovanjem med večjim in manjšim, katero ti ne gre? A ti razumeš da z zajemanjem vode s cedilom ne moreš natankat posode ali ne?
Najbrž ne. Pa še en cajt ne boš.

Pfizer je kupil podjetje, ki se specializira za turbo raka:

 www.biotecnika.org
www.biotecnika.org

Shocking Revelation: Pfizer's $43 Billion Gamble and the Truth About Turbo Cancers Linked to mRNA Vaccines
Pfizer's $43 Billion Bet: Understanding the Rise of 'Turbo Cancers' Pfizer Cancer Research. Cancer Vaccinations.
AndY, prideš plačat svoj dolg?
Je to zdravilo med pandemijo covid-19 povzročilo na tisoče smrti?
Glede na študijo francoskih raziskovalcev je skoraj 17 tisoč ljudi v več državah po svetu morda umrlo zaradi jemanja hidroksiklorokina med prvim valom covid-19. Gre za zdravilo za malarijo, katerega jemanje je spodbujal tudi tedanji ameriški predsednik Donald Trump.siol.net
To zdravilo naj bi imelo neželene učinke, kot so motnje srčnega ritma.
ki so morda umrli zaradi uživanja hidroksiklorokina.
Lahko prilepis se bolj zavajajoc clanek?
opozorilo o nevarnosti mRNA cepiva je bilo izdano leta 2007.
Torej: vsi predsedniki držav, predsedniki vlad, zdravstveni ministri in podobni kader, ki edini ima kaj vpliva na delovanje in dobiček farmacevtskih družb, je bil smeje, pred kamero, cepljen z mRNA cepivom ker so ocenili da so "koristi večje od tveganja".
Ali to, ali pa cepljenje z 0,9% raztopino natrijevega klorida, oziroma s slano vodo.
Globoko verjamem da so izbrali mRNA, zaradi svojega zdravja, zdravja svoje družine, in da se gospodarstvo in družba na splošno čimprej pobere.

 rumble.com
rumble.com
Torej: vsi predsedniki držav, predsedniki vlad, zdravstveni ministri in podobni kader, ki edini ima kaj vpliva na delovanje in dobiček farmacevtskih družb, je bil smeje, pred kamero, cepljen z mRNA cepivom ker so ocenili da so "koristi večje od tveganja".
Ali to, ali pa cepljenje z 0,9% raztopino natrijevega klorida, oziroma s slano vodo.
Globoko verjamem da so izbrali mRNA, zaradi svojega zdravja, zdravja svoje družine, in da se gospodarstvo in družba na splošno čimprej pobere.

DNA and Florida
Tallahassee, Fla. State Surgeon General Dr. Joseph A. Ladapo https://twitter.com/FLSurgeonGen https://content.govdelivery.com/accounts/FLDOH/bulletins/3816863 https://www.floridahealth.gov/newsroom/20
Ni potrebe, si jih že ti dovolj.Lahko prilepis se bolj zavajajoc clanek?
Presezne smrti po dobrem letu prihajajo v main stream medije:

 thehill.com
thehill.com
Life insurers have been consistently sounding the alarm over these unexpected or, “excess,” deaths, which claimed 158,000 more Americans in the first nine months of 2023 than in the same period in 2019. That exceeds America’s combined losses from every war since Vietnam.
Actuarial reports — used by insurers to inform decisions — show deaths occurring disproportionately among young working-age people. Nonetheless, America’s chief health manager, the Centers for Disease Control and Prevention, opted in September to archive its excess deaths webpage with a note stating, “these datasets will no longer be updated.”
Unlike in the pandemic’s early phase, these deaths are not primarily among the old. For people 65 and over, deaths in the second quarter of 2023 were 6 percent below the pre-pandemic norm, according to a new report from the Society of Actuaries. Mortality was 26 percent higher among insured 35-to-44-year-olds, and 19 percent higher for 25-to-34-year-olds, continuing a death spike that peaked in the third quarter of 2021 at a staggering 101 percent and 79 percent above normal, respectively.

This is bigger than COVID: Why are so many Americans dying early?
Why is the traditionally healthiest sector of our society — young, employed, insured workers — dying at such rates?
This is bigger than COVID: Why are so many Americans dying early?
People are dying in abnormally high numbers even now and long since COVID-19 waned. Yet public health agencies and medical societies are silent.Life insurers have been consistently sounding the alarm over these unexpected or, “excess,” deaths, which claimed 158,000 more Americans in the first nine months of 2023 than in the same period in 2019. That exceeds America’s combined losses from every war since Vietnam.
Actuarial reports — used by insurers to inform decisions — show deaths occurring disproportionately among young working-age people. Nonetheless, America’s chief health manager, the Centers for Disease Control and Prevention, opted in September to archive its excess deaths webpage with a note stating, “these datasets will no longer be updated.”
Unlike in the pandemic’s early phase, these deaths are not primarily among the old. For people 65 and over, deaths in the second quarter of 2023 were 6 percent below the pre-pandemic norm, according to a new report from the Society of Actuaries. Mortality was 26 percent higher among insured 35-to-44-year-olds, and 19 percent higher for 25-to-34-year-olds, continuing a death spike that peaked in the third quarter of 2021 at a staggering 101 percent and 79 percent above normal, respectively.
Ivermectin tudi po racunalniskih simulacijah (in-silico) ucinkovito deluje proti Sars-Cov2 virusu:
 www.ncbi.nlm.nih.gov
www.ncbi.nlm.nih.gov
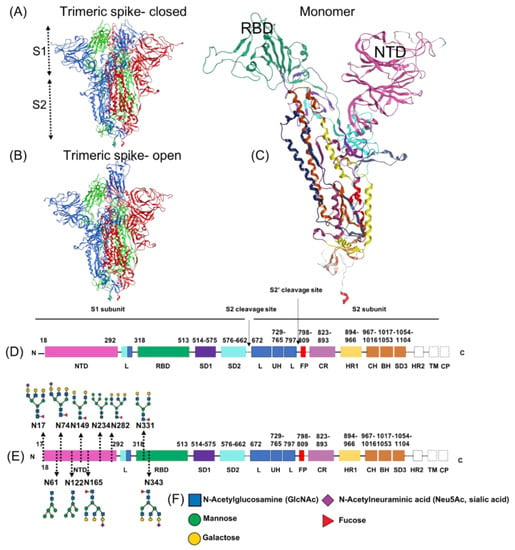
 www.mdpi.com
www.mdpi.com
Among all the targets, ivermectin has the highest affinity to the α7nAChr receptor. Protein–protein docking results reveal a potential direct binding of the SARS-CoV-2 spike-1 protein to α7nAChr, suggesting that this ubiquitous cholinergic receptor may mediate SARS-CoV-2 entry into cells, shedding light on multiple aspects of SARS-CoV-2 infection pathophysiology (i.e., the loss of smell and taste, the cytokine storm and impairment of the endothelium-dependent acetylcholine-induced vasodilation). In this context, the high affinity of ivermectin and related compounds to α7nAChr may both prevent viral entry and potentiate the activation of the cholinergic pathway and attenuate SARS-CoV-2-induced parasympathetic dysregulation by restoring the function of these receptors. Our preliminary results warrant further in vitro and in vivo testing of the 15 test compounds, in particular ivermectin, an available and safe drug, against SARS-CoV-2, with the hope of containing the virus and limiting its morbidity.
18:33
An in-silico analysis of ivermectin interaction with potential SARS-CoV-2 targets and host nuclear importin α - PMC
Ivermectin (IVM) is a broad-spectrum antiparasitic agent, having inhibitory potential against wide range of viral infections. It has also been found to hamper SARS-CoV-2 replication in vitro, and its precise mechanism of action against SARS-CoV-2 is ...
Conclusion
Ivermectin, a broad-spectrum antiparasitic agent, has been found to inhibit SARS-CoV-2 replication in vitro. In this study, 15 potential COVID-19 targets were used to underscore IVM affinity by computational techniques employing molecular docking, MM/GBSA computation and molecular dynamics simulation. IMPα protein was also included in this study owing to the reported interaction of IVM with host importin α, resulting in the prevention of nuclear localization signal recognition. Estimation of MM/GBSA based binding energy revealed that Nsp9 possesses highest affinity among COVID-19 targets. Strengths of IVM complex with Nsp9 and IMPα was ascertained by the evaluation of RMSD and RMSF plots obtained after molecular dynamics simulation of 100 ns. Both hydrophobic as well as hydrophilic interactions were identified in anchoring the IVM inside the binding site of Nsp9 as well as major binding groove of the IMPα. Intermolecular interaction profile of IVM disclosed in the current study is expected to assist experimental studies and designing of COVID-19 drugs.
In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds
Some clinical studies have indicated activity of ivermectin, a macrocyclic lactone, against COVID-19, but a biological mechanism initially proposed for this anti-viral effect is not applicable at physiological concentrations. This in silico investigation explores potential modes of action of...
Conclusions
In the present study, a computational investigation including molecular docking was conducted to explore the potential bindings of ivermectin and 14 similar compounds to three targets of interest (the spike, CD147 and α7nAChr) that are relevant for drug activity against COVID-19. Strong or moderate affinity bindings were found for ivermectin to multiple sites on the spike protein, CD147 and α7nAChr, which could provide effective competitive bindings to all variants of the virus. According to our calculations, ivermectin binds strongly to a glycosylation binding site (site 10: N61) of the spike protein S1-NTD in the open position and to several other sites on S1 NTD and RBD. We also examined the potential bindings of ivermectin to CD147. Ivermectin was found to bind to site 5, which is located in domain A of the CD147 protein, and to other sites on CD147, indicating that ivermectin might limit glycan bindings of SARS-CoV-2 at the host cell end as well.Among all the targets, ivermectin has the highest affinity to the α7nAChr receptor. Protein–protein docking results reveal a potential direct binding of the SARS-CoV-2 spike-1 protein to α7nAChr, suggesting that this ubiquitous cholinergic receptor may mediate SARS-CoV-2 entry into cells, shedding light on multiple aspects of SARS-CoV-2 infection pathophysiology (i.e., the loss of smell and taste, the cytokine storm and impairment of the endothelium-dependent acetylcholine-induced vasodilation). In this context, the high affinity of ivermectin and related compounds to α7nAChr may both prevent viral entry and potentiate the activation of the cholinergic pathway and attenuate SARS-CoV-2-induced parasympathetic dysregulation by restoring the function of these receptors. Our preliminary results warrant further in vitro and in vivo testing of the 15 test compounds, in particular ivermectin, an available and safe drug, against SARS-CoV-2, with the hope of containing the virus and limiting its morbidity.
Conclusion
Developing an effective therapeutic against COVID-19 is currently the utmost interest to the scientific communities. The present study depicts comparative binding efficacy of a promising FDA-approved drug, ivermectin, against major pathogenic proteins of SARS-CoV-2 and their human counterparts involved in host–pathogen interaction. Herein, our in silico data have indicated that ivermectin efficiently utilizes viral spike protein, main protease, replicase and human TMPRSS2 receptors as the most possible targets for executing its antiviral efficiency. Therefore, ivermectin exploits protein targets from both virus and human, which could be the reason behind its excellent in vitro efficacy against SARS-CoV-2 as reported by Caly et al. [13]. Ivermectin B1b isomers have been found to be the more efficacious molecule out of the two homologs. Intriguingly, comparison of the in silico efficiency of ivermectin with currently used anticorona drugs, such as hydroxychloroquine and remdesivir, indicated toward the potential of ivermectin to target the major pathogenic proteins of SARS-CoV-2. Ivermectin is a popular antiparasitic drug and is also safe in children, younger adults, pregnant and lactating ladies. Development of pulmonary delivery of ivermectin through synthesis of better ivermectin formulation has been reported recently and this is expected to shorten the treatment duration and lead to better outcomes [33]. It is noteworthy to mention that many anti-SARS-CoV-2s are now being tested for their efficacy in shaping the immune response of humans, through targeting the cell surface as well as intracellular toll-like receptors [34,35]. In this context, ivermectin could be an effective option as well. Considering all these facts, the present study explores the therapeutic targets of ivermectin against SARS-CoV-2 and enlightens the possibility of using this drug in COVID-19 clinical trials shortly.18:33
Mah, kovid je tako že lanski sneg.
Niso pa posledice Covid cepiv.
Nekoc ste se mi režali, zdaj so dolgi beli strdki v zilah umrlih jasno dejstvo:
Smešno vam je toliko časa, dokler vam ne bo več. Takrat bo pa nam ostalim smešno.Režimo se ti še danes

Nekoc ste se mi režali, zdaj so dolgi beli strdki v zilah umrlih jasno dejstvo:
na Youtube so omejili dostop, tukaj je še dostopen:

What are these
je pa tukaj še daljše pojasnilo, ne samo glede teh strdkov, tudi glede okoliščin, npr. koliko denarja so zdravniki dobili od vsake vakcine če so prepričali 70% svojih pacientov da naj se gredo cepit (moj dohtar se je dobro znašel: na vhodna vrata nalepil velik napis "v tej ambulanti priporočamo cepljenje proti covid") ; zakaj število presežnih smrti posamezniku mogoče ni takoj očitno; da bi patologi (ali kako se reče tistim ki izvajajo obdukcije?) želeli javno predstaviti kar najdejo pri svojem delu, pa za to nimajo možnosti, ali pa jim to ni dovoljeno; dovolj dobro razumljivo z osnovnošolskim znanjem angleščine, tudi na 1.25-x hitrost predvajanja; kakšno je stanje v Sloveniji glede tega?
Podobne teme
- Odgovori
- 163
- Ogledi
- 57.119


